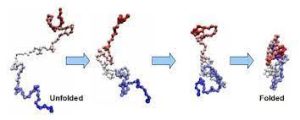
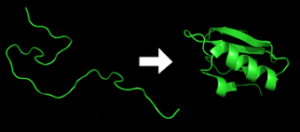
 Protein folding is the physical process where a protein chain is translated into its native three-dimensional structure.
Protein folding is the physical process where a protein chain is translated into its native three-dimensional structure.
Typically a “folded” conformation, allows the protein to become biologically functional.
Polypeptides fold into its characteristic three-dimensional structure from a random coil.
Proteins exists first as an unfolded polypeptide or random coil.
Proteins are translated from a sequence of mRNA into a linear chain of amino acids, and at this stage, the polypeptide lacks any stable three-dimensional structure.
As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure.
Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein resulting three-dimensional structure is determined by the amino-acid sequence or primary structure.
Failure to fold into a native three-dimensional structure generally produces inactive proteins.
In some instances, misfolded proteins have modified/toxic functionality.
Several neurodegenerative diseases result from the accumulation of amyloid fibrils formed by misfolded proteins.
Infectious varieties of misfolded proteins are known as prions.
Many allergies are caused by the incorrect folding of some proteins:the immune system does not produce the antibodies for certain protein structures.
Denaturation of proteins refers to the transition from a folded to an unfolded state: occurs in cooking, burns, proteinopathies, and other contexts.
Protein folding refers to the process by which a newly synthesized protein molecule acquires its three-dimensional structure, known as its native conformation.
Protein folding process is crucial for the proper functioning of proteins in cells.
The primary structure of a protein is the linear sequence of amino acids joined together by peptide bonds, and is encoded by the DNA in a gene.
The specific order of amino acids determines how the protein will fold into its functional shape.
Protein folding occurs via specific pathways, often guided by the principle of lowest energy state.
The folding process is influenced by amino acid sequence, pH, temperature, and the presence of other molecules, known as molecular chaperones, which assist in the folding process.
The folding process initially leads to the formation of regular structural elements within the protein, such as alpha-helices and beta-sheets.
Alpha-helices and beta-sheets structures are stabilized by hydrogen bonds between the backbone atoms of the amino acids.
Further folding involves three-dimensional arrangement of the protein molecule, including the interaction and positioning of different secondary structures.
Forces contribute to the stabilization of the tertiary structure: hydrogen bonds, disulfide bonds, electrostatic interactions, hydrophobic interactions, and van der Waals forces.
Some proteins consisting of multiple individual subunits assemble to form a larger, functional protein complex, the quaternary structure: hemoglobin, which consists of four subunits, and antibodies, which often have two heavy chains and two light chains.
The correct folding of proteins is essential to biological function, determining its ability to bind to other molecules, catalyze chemical reactions, transport substances, and carry out other biochemical activities necessary for cellular processes.
Proteins may misfold, failing to attain their proper native conformation.
Such misfolded proteins can aggregate, forming clumps associated with various diseases, including neurodegenerative disorders such as Alzheimer’s, Parkinson’s, and Huntington’s disease.
Computational methods, X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, are used to study and predict protein folding.
When studied outside the cell, the slowest folding proteins require many minutes or hours to fold, primarily due to proline isomerization, passing through a number of intermediate states before the process is complete.
Very small single-domain proteins with lengths of up to a hundred amino acids typically fold in a single step, in milliseconds.
The fastest known protein folding reactions are complete within a few microseconds.
The folding time of a protein depends on its size, contact order, and circuit topology.
The primary structure of a protein is its linear amino-acid sequence, which determines its native conformation.
The specific amino acid residues and their position in the polypeptide chain are the determining factors for which portions of the protein fold closely together and form its three-dimensional conformation.
The amino acid composition is not as important as the sequence for folding.
The essential fact of folding, however, remains that the amino acid sequence of each protein contains the information that specifies both the native structure and the pathway to attain that state.
Protein conformations differ based on environmental factors as well; similar proteins fold differently based on where they are found.
Formation of a secondary structure is the first step in the folding process that a protein takes to assume its native structure.
Alpha helices and beta sheets are secondary structures that fold rapidly because they are stabilized by intramolecular hydrogen bonds.
α-helices are formed by hydrogen bonding of the backbone to form a spiral shape .
The β pleated sheet is a structure that forms with the backbone bending over itself to form the hydrogen bonds.
The α-Helices and β-Sheets have a hydrophilic and a hydrophobic portion, which helps in forming a tertiary structure of a protein
in which folding occurs so that the hydrophilic sides are facing the aqueous environment surrounding the protein and the hydrophobic sides are facing the hydrophobic core of the protein.
Secondary structure hierarchically gives way to tertiary structure formation.
Tertiary structures are formed and stabilized by the hydrophobic interaction.
Tertiary structure of a protein involves a single polypeptide chain; however, additional interactions of folded polypeptide chains give rise to quaternary structure formation.
Tertiary structure may give way to the formation of quaternary structure in some proteins, which usually involves the multiple polypeptide chains that interact to form a fully functional quaternary protein.
Folding is a spontaneous process guided by hydrophobic interactions, formation of intramolecular hydrogen bonds, van der Waals forces.
The folding process depends on the solvent: water or lipid bilayer,
concentration of salts, the pH, the temperature, the possible presence of cofactors and of molecular chaperones.
Proteins have limitations on their folding abilities, restricted bending angles or conformations that are possible.
Molecular chaperones are proteins that aid in the correct folding of other proteins in vivo.
Chaperones exist in all cellular compartments and interact with the polypeptide chain in order to allow the native three-dimensional conformation of the protein to form.
Molecular chaperones operate by binding to stabilize an unstable structure of a protein in its folding pathway.
Chaperones work by preventing incorrect folding conformations, by reducing possible unwanted aggregations of the polypeptide chain that might otherwise slow down the search for the proper intermediate and they provide a more efficient pathway for the polypeptide chain to assume the correct conformations.
Chaperones are shown to be involved in various roles such as protein transport, degradation, and even allow denatured proteins exposed to certain external denaturant factors an opportunity to refold into their correct native structures.
A fully denatured protein lacks both tertiary and secondary structure.
Cells sometimes protect their proteins against the denaturing influence of heat with enzymes known as heat shock proteins , which are a type of chaperone.
Heat shock proteins assist other proteins both in folding and in remaining folded.
Chaperones increase in concentration during times of cellular stress and help the proper folding of emerging proteins as well as denatured or misfolded ones.
Under some conditions proteins will not fold into their biochemically functional forms.
Temperatures above or below the range that cells tend to live in will cause thermally unstable proteins to unfold or denature.
Some proteins have multiple native structures, and change their fold based on some external factors.
A misfolded protein cannot achieve its normal native state, and may be
due to mutations in the amino acid sequence or a disruption of the normal folding process by external factors.
A misfolded protein typically contains β-sheets that are organized in a supramolecular arrangement known as a cross-β structure.
These β-sheet-rich assemblies are very stable, very insoluble, and generally resistant to proteolysis.
β-sheet stability of these fibrillar assemblies is caused by extensive interactions between the protein monomers, formed by backbone hydrogen bonds between their β-strands.
The misfolding of proteins can trigger the further misfolding and accumulation of other proteins into aggregates or oligomers.
The increased levels of aggregated proteins in the cell leads to formation of amyloid-like structures which can cause degenerative disorders and cell death.
The amyloids are fibrillary structures with intermolecular hydrogen bonds which are highly insoluble and made from converted protein aggregates.
The proteasome pathway may not be efficient enough to degrade the misfolded proteins prior to aggregation.
Misfolded proteins can interact with one another forming aggregates and gain toxicity through intermolecular interactions.
Aggregated proteins are associated with prion-related illnesses such as Creutzfeldt–Jakob disease, bovine spongiform encephalopathy (mad cow disease), amyloid-related illnesses such as Alzheimer’s disease and familial amyloid cardiomyopathy or polyneuropathy, as well as intracellular aggregation diseases such as Huntington’s and Parkinson’s disease.
Age onset degenerative diseases are associated with the aggregation of misfolded proteins into insoluble, extracellular aggregates and/or intracellular inclusions including cross-β amyloid fibrils.
It is not for sure whether the aggregates are the cause or merely a reflection of the loss of protein homeostasis, which is the balance between synthesis, folding, aggregation and protein turnover.
Tafamidis or Vyndaqel is a a kinetic stabilizer of tetrameric transthyretin for the treatment of transthyretin amyloid diseases, suggesting that the process of amyloid fibril formation, and not the fibrils themselves, that causes the degeneration of post-mitotic tissue in human amyloid diseases.
Misfolding and excessive degradation instead of folding and function leads to a number of proteopathy diseases such as antitrypsin-associated emphysema, cystic fibrosis and the lysosomal storage diseases, where loss of function is the origin of the disorder.
While protein replacement therapy has been used to correct the latter disorders.
The use of pharmaceutical chaperones to fold mutated proteins to render them functional, is in investigational stages.
X-ray crystallography is one of the methods for attempting to decipher the three dimensional configuration of a folded protein,as is fluorescence spectroscopy.
Circular dichroism spectroscopy measures the absorption of circularly polarized light.
Protein structures such as alpha helices and beta sheets are chiral, and thus absorb such light, and the absorption of this light acts as a marker of the degree of foldedness of the protein ensemble.
Crcular dichroism of proteins provides powerful means for determining protein conformations in solution even for very large protein molecules.
Protein nuclear magnetic resonance (NMR) is able to collect protein structural data by inducing a magnet field through samples of concentrated protein.
Dual polarisation interferometry is a surface-based technique for measuring the optical properties of molecular layers.
Computational studies of protein folding includes the prediction of protein stability, kinetics, and structure.
The folding landscape allows the protein to fold to the native state through any of a large number of pathways and intermediates, rather than being restricted to a single mechanism.
As a protein begins to fold and assume its various conformations, it always seeks a more thermodynamically favorable structure than before and thus continues through the energy funnel.
Protein misfolding occurs during different biochemical processes and may lead to the development of diseases such as cancer, which is characterized by genetic instability.
The cancer microenvironment exposes malignant cells to a variety of stressful conditions that may further promote protein misfolding.
